Abstract
Introduction
Extramedullary disease (EMD) is a less frequent manifestation of multiple myeloma (MM), where MM plasma cells become independent of the bone marrow (BM) microenvironment and infiltrate other tissues and organs. The incidence of EMD is increasing and is associated with worse prognosis and drug resistance. The specific and efficient treatment is lacking. Therefore, a better understanding of EMD pathogenesis is desperately needed.
Aims
To identify biological pathways leading to EMD development and to evaluate therapeutic targets in EMD plasma cells with further focus on EMD tumor microenvironment to reveal presence of effector immune cells that are crucial for immunotherapy.
Methods
To identify EMD specific genes, FACS/MACS sorted aberrant plasma cells were collected from: i) fresh 11 EMD relapse tumors for which we had ii) 7 corresponding cryopreserved paired BM samples from the time of MM diagnosis (NDMM), iii) 9 unpaired fresh NDMM without EMD confirmed by PET-CT and iv) 6 unpaired fresh relapsed MM (RRMM). For library preparation, we used total RNA with rRNA depletion protocol and Illumina sequencing. Residual rRNA was filtered out by SortMeRNA. Differential expression analysis was performed using Salmon for read mapping and quantification and Deseq2 package. For single-cell RNAseq we used 10x Genomics technology for sequencing and CellRanger and Seurat for data processing and analysis.
Results
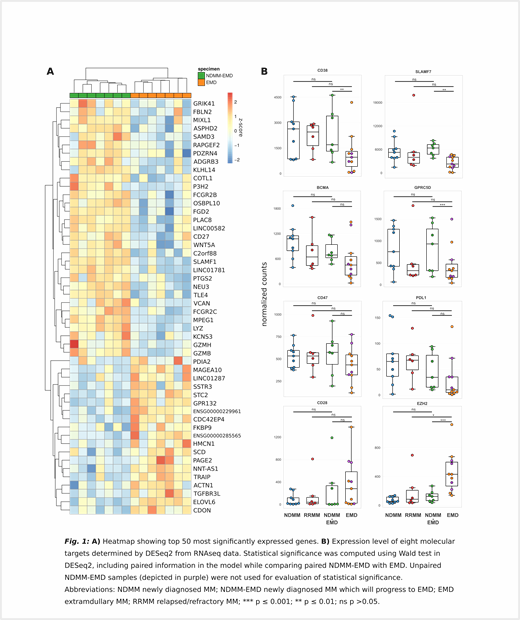
To better understand the aggressive nature of EMD, we have analyzed bulk RNA samples (7 EMD samples plus 7 corresponding cryopreserved paired BM samples from the time of MM diagnosis). Our preliminary analysis revealed a unique EMD profile (Fig 1A) with 423 up-regulated and 421 down-regulated genes in EMD samples (adjusted p-value < 0.1; absolute fold change > 1.5), with G2M checkpoint proteins being the most enriched hallmark pathways pointing to higher proliferation of EMD cells. EMD down-regulated genes mainly belong to genes of the adaptive immune response which together with lower immunoglobulin production suggest loss of mature plasma cell function.
Among the top genes uniquely overexpressed in EMD (versus RRMM or NDMM) were SCD and ELOVL6 that regulate crucial steps in unsaturated fatty acids synthesis. Also their transcription factor SREBF1 was significantly up-regulated. The importance of these genes in EMD pathogenesis can be supported by the involvement of SREBP1 in stem cell differentiation and mediation of bortezomib resistance by ELOVL6 (Yi et al. 2018, Lipchick et al. 2021). Our dataset also revealed several deregulated lncRNA in EMD compared to NDMM. MALAT1 was highly expressed, however, we did not confirm results by Handa et al. 2017 showing lncRNA MALAT1 as upregulated in EMD.
Furthermore, we aimed to evaluate expression of known immunotherapy MM targets being currently in use or under investigation. We compared the information about expression level in EMD vs paired NDMM, with unpaired NDMM without EMD lesion confirmed by PET/CT, and with RRMM. The analysis revealed a decrease in the expression of several antigens commonly used in anti-MM immunotherapy (e.g. CD38, SLAMF7, BCMA or PDL1) on EMD PCs (Fig 1B). Intriguingly, our data show EMD specific elevated expression of EZH2 gene being promising target in preclinical MM investigation which can prove efficient especially for the aggressive MM stage - EMD.
Effective immunotherapy depends on the presence of effector immune cells. Therefore, we have evaluated immune cell types and their proportion in EMD tumors. Using flow cytometry we identified T and NK cells as the only immune cell subsets present in EMD tumors (median 0.9% and 0.5%, respectively). Single-cell RNAseq analysis of two EMD samples supported these findings.
Conclusions
Here, we present up to our knowledge the worldwide largest cohort of 11 EMD samples (including 7 longitudinal pre-EMD/EMD samples) analysed using RNAseq with focus on biological pathways and dysregulation of particular genes leading to EMD development. Drop of expression of several known drug targets may suggest limited efficacy of the modern treatment in EMD as already presented by Jelinek et al., 2021. Importantly, we are also providing the initial insight into the microenvironment (including single-cell RNA analysis) of EMD tumors, where we detected presence of T cell and NK cells in very limited numbers.
Hajek: Takeda: Consultancy, Honoraria, Membership on an entity's Board of Directors or advisory committees, Research Funding; Celgene: Consultancy, Honoraria, Research Funding; BMS: Consultancy, Honoraria, Research Funding; AbbVie: Consultancy, Honoraria; Novartis: Consultancy, Research Funding; Pharma MAR: Consultancy, Honoraria; Janssen: Consultancy, Honoraria, Research Funding; Amgen: Consultancy, Honoraria, Membership on an entity's Board of Directors or advisory committees, Research Funding.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal